clisops regridding functionalities - powered by xesmf
The regridding functionalities of clisops consist of the regridding operator/function regrid in clisops.ops, allowing one-line remapping of xarray.Datasets or xarray.DataArrays, while orchestrating the use of classes and functions in clisops.core: - the Grid and Weights classes, to check and pre-process input as well as output grids and to generate the remapping weights - a regrid function, performing the remapping by applying the generated weights on the input
data
For the weight generation and the regridding, the xESMF Regridder class is used, which itself allows an easy application of many of the remapping functionalities of ESMF/ESMPy.
[1]:
# Imports
%matplotlib inline
import matplotlib.pyplot as plt
import cartopy.crs as ccrs
import psyplot.project as psy
import numpy as np
import xarray as xr
import cf_xarray as cfxr
from pathlib import Path
from git import Repo
# Set required environment variable for ESMPy
import os
os.environ['ESMFMKFILE'] = str(Path(os.__file__).parent.parent / 'esmf.mk')
import xesmf as xe
import clisops as cl # atm. the regrid-main-martin branch of clisops
import clisops.ops as clops
import clisops.core as clore
from clisops.utils import dataset_utils
from roocs_grids import get_grid_file, grid_dict, grid_annotations
print(f"Using xarray in version {xr.__version__}")
print(f"Using cf_xarray in version {cfxr.__version__}")
print(f"Using xESMF in version {xe.__version__}")
print(f"Using clisops in version {cl.__version__}")
xr.set_options(display_style='html')
## Turn off warnings?
import warnings
warnings.simplefilter("ignore")
Matplotlib is building the font cache; this may take a moment.
ERROR 1: PROJ: proj_create_from_database: Open of /home/docs/checkouts/readthedocs.org/user_builds/clisops/conda/stable/share/proj failed
Using xarray in version 2023.2.0
Using cf_xarray in version 0.8.9
Using xESMF in version 0.8.2
Using clisops in version 0.13.0
[2]:
# Initialize test data
# Initialize mini-esgf-data
MINIESGF_URL="https://github.com/roocs/mini-esgf-data"
branch = "master"
MINIESGF = Path(Path.home(),".mini-esgf-data", branch)
# Retrieve mini-esgf test data
if not os.path.isdir(MINIESGF):
repo = Repo.clone_from(MINIESGF_URL, MINIESGF)
repo.git.checkout(branch)
else:
repo = Repo(MINIESGF)
repo.git.checkout(branch)
repo.remotes[0].pull()
MINIESGF=Path(MINIESGF,"test_data")
clisops.ops.regrid
One-line remapping with clisops.ops.regrid
def regrid(
ds: Union[xr.Dataset, str, Path],
*,
method: Optional[str] = "nearest_s2d",
adaptive_masking_threshold: Optional[Union[int, float]] = 0.5,
grid: Optional[
Union[xr.Dataset, xr.DataArray, int, float, tuple, str]
] = "adaptive",
output_dir: Optional[Union[str, Path]] = None,
output_type: Optional[str] = "netcdf",
split_method: Optional[str] = "time:auto",
file_namer: Optional[str] = "standard",
keep_attrs: Optional[Union[bool, str]] = True,
) -> List[Union[xr.Dataset, str]]
The different options for the method, grid and adaptive_masking_threshold parameters are described in below sections:
Remap a global xarray.Dataset to a global 2.5 degree grid using the bilinear method
Load the dataset
[3]:
ds_vert_path = Path(MINIESGF, "badc/cmip6/data/CMIP6/CMIP/MPI-M/MPI-ESM1-2-LR/historical/r1i1p1f1/AERmon/"
"o3/gn/v20190710/o3_AERmon_MPI-ESM1-2-LR_historical_r1i1p1f1_gn_185001.nc")
ds_vert = xr.open_dataset(ds_vert_path)
ds_vert
[3]:
<xarray.Dataset>
Dimensions: (time: 1, bnds: 2, lon: 192, lat: 96, lev: 3)
Coordinates:
* time (time) datetime64[ns] 1850-01-16T12:00:00
* lon (lon) float64 0.0 1.875 3.75 5.625 ... 352.5 354.4 356.2 358.1
* lat (lat) float64 -88.57 -86.72 -84.86 -83.0 ... 84.86 86.72 88.57
* lev (lev) float64 0.9961 0.9826 0.959
Dimensions without coordinates: bnds
Data variables:
time_bnds (time, bnds) datetime64[ns] ...
lon_bnds (lon, bnds) float64 ...
lat_bnds (lat, bnds) float64 ...
lev_bnds (lev, bnds) float64 ...
ap (lev) float64 ...
b (lev) float64 ...
ap_bnds (lev, bnds) float64 ...
b_bnds (lev, bnds) float64 ...
ps (time, lat, lon) float32 ...
o3 (time, lev, lat, lon) float32 ...
Attributes: (12/49)
CDI: Climate Data Interface version 1.9.8 (https://mpi...
history: Tue Oct 26 11:48:46 2021: cdo sellevidx,1/3 -selt...
source: MPI-ESM1.2-LR (2017): \naerosol: none, prescribed...
institution: Max Planck Institute for Meteorology
Conventions: CF-1.7 CMIP-6.2
activity_id: CMIP
... ...
variable_id: o3
variant_label: r1i1p1f1
license: CMIP6 model data produced by MPI-M is licensed un...
cmor_version: 3.5.0
tracking_id: hdl:21.14100/52bee89e-e989-4005-8663-e60210d421d5
CDO: Climate Data Operators version 1.9.8 (https://mpi...Take a look at the grid
[4]:
# Create 2D coordinate variables
lon,lat = np.meshgrid(ds_vert["lon"].data, ds_vert["lat"].data)
# Plot
plt.figure(figsize=(8,5))
plt.scatter(lon[::3, ::3], lat[::3, ::3], s=0.5)
plt.xlabel('lon')
plt.ylabel('lat')
[4]:
Text(0, 0.5, 'lat')
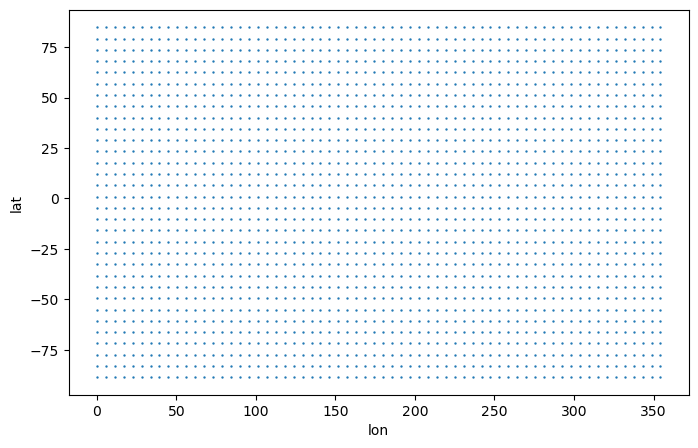
Remap to global 2.5 degree grid with the bilinear method
[5]:
ds_remap = clops.regrid(ds_vert, method="bilinear", grid="2pt5deg", output_type="xarray")[0]
ds_remap
[5]:
<xarray.Dataset>
Dimensions: (lat: 72, lon: 144, bnds: 2, time: 1, lev: 3)
Coordinates:
* lat (lat) float64 -88.75 -86.25 -83.75 -81.25 ... 83.75 86.25 88.75
* lon (lon) float64 1.25 3.75 6.25 8.75 ... 351.2 353.8 356.2 358.8
lat_bnds (lat, bnds) float64 -90.0 -87.5 -87.5 -85.0 ... 87.5 87.5 90.0
lon_bnds (lon, bnds) float64 0.0 2.5 2.5 5.0 ... 355.0 357.5 357.5 360.0
* time (time) datetime64[ns] 1850-01-16T12:00:00
* lev (lev) float64 0.9961 0.9826 0.959
ap (lev) float64 0.0 0.0 36.03
b (lev) float64 0.9961 0.9826 0.9586
time_bnds (time, bnds) datetime64[ns] 1850-01-01 1850-02-01
ap_bnds (lev, bnds) float64 0.0 0.0 0.0 0.0 0.0 72.06
b_bnds (lev, bnds) float64 1.0 0.9923 0.9923 0.973 0.973 0.9442
lev_bnds (lev, bnds) float64 1.0 0.9923 0.9923 0.973 0.973 0.9449
Dimensions without coordinates: bnds
Data variables:
ps (time, lat, lon) float32 7.025e+04 7.018e+04 ... 1.01e+05
o3 (time, lev, lat, lon) float32 1.671e-08 1.669e-08 ... 2.321e-08
Attributes: (12/54)
CDI: Climate Data Interface version 1.9.8 (https...
history: Tue Oct 26 11:48:46 2021: cdo sellevidx,1/3...
source: MPI-ESM1.2-LR (2017): \naerosol: none, pres...
institution: Max Planck Institute for Meteorology
Conventions: CF-1.7 CMIP-6.2
activity_id: CMIP
... ...
grid_original: gn
grid_label_original: gn
nominal_resolution_original: 250 km
regrid_operation: bilinear_96x192_72x144_peri
regrid_tool: xESMF_v0.8.2
regrid_weights_uid: 20d38455170b1db562edb4f5e06d3cc6_d4ebf55323...Plot the remapped data next to the source data
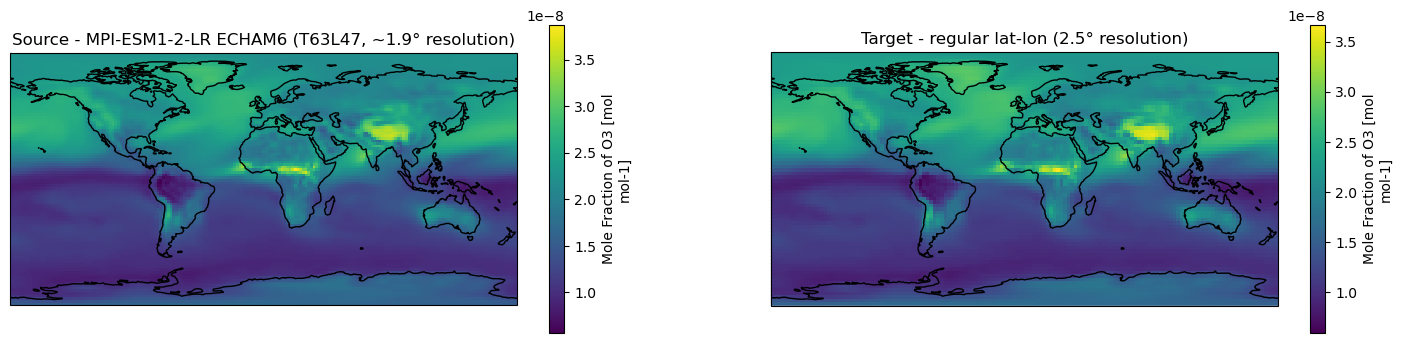
[6]:
fig, axes = plt.subplots(ncols=2, figsize=(18,4), subplot_kw={'projection': ccrs.PlateCarree()})
for ax in axes:
ax.coastlines()
# Source data
ds_vert.o3.isel(time=0, lev=0).plot.pcolormesh(ax=axes[0], x="lon", y="lat", shading="auto")
axes[0].title.set_text("Source - MPI-ESM1-2-LR ECHAM6 (T63L47, ~1.9° resolution)")
# Remapped data
ds_remap.o3.isel(time=0, lev=0).plot.pcolormesh(ax=axes[1], x="lon", y="lat", shading="auto")
axes[1].title.set_text("Target - regular lat-lon (2.5° resolution)")
Remap regional xarray.Dataset to a regional grid of adaptive resolution using the bilinear method
Adaptive resolution means, that the regular lat-lon target grid will have approximately the same resolution as the source grid.
Load the dataset
[7]:
ds_cordex_path = Path(MINIESGF, "pool/data/CORDEX/data/cordex/output/EUR-22/GERICS/MPI-M-MPI-ESM-LR/"
"rcp85/r1i1p1/GERICS-REMO2015/v1/mon/tas/v20191029/"
"tas_EUR-22_MPI-M-MPI-ESM-LR_rcp85_r1i1p1_GERICS-REMO2015_v1_mon_202101.nc")
ds_cordex = xr.open_dataset(ds_cordex_path)
ds_cordex
[7]:
<xarray.Dataset>
Dimensions: (rlat: 201, rlon: 225, vertices: 4, time: 1,
bnds: 2)
Coordinates:
height float64 ...
lat (rlat, rlon) float32 ...
lon (rlat, rlon) float32 ...
* rlat (rlat) float64 -22.88 -22.66 ... 20.9 21.12
* rlon (rlon) float64 -29.86 -29.64 ... 19.2 19.42
* time (time) datetime64[ns] 2021-01-16T12:00:00
Dimensions without coordinates: vertices, bnds
Data variables:
lat_vertices (rlat, rlon, vertices) float32 ...
lon_vertices (rlat, rlon, vertices) float32 ...
rotated_latitude_longitude int32 ...
tas (time, rlat, rlon) float32 ...
time_bnds (time, bnds) datetime64[ns] ...
Attributes: (12/35)
institution: Helmholtz-Zentrum Geesthacht, Climate Ser...
institute_id: GERICS
experiment_id: rcp85
source: GERICS-REMO2015
model_id: GERICS-REMO2015
forcing: N/A
... ...
parent_experiment: N/A
modeling_realm: atmos
realization: 1
cmor_version: 2.9.1
tracking_id: hdl:21.14103/77bb3489-7951-4f31-bf63-3022...
NCO: netCDF Operators version 4.7.5 (Homepage ...Take a look at the grid
[8]:
plt.figure(figsize=(8,5))
plt.scatter(ds_cordex['lon'][::4, ::4], ds_cordex['lat'][::4, ::4], s=0.1)
plt.xlabel('lon')
plt.ylabel('lat')
[8]:
Text(0, 0.5, 'lat')
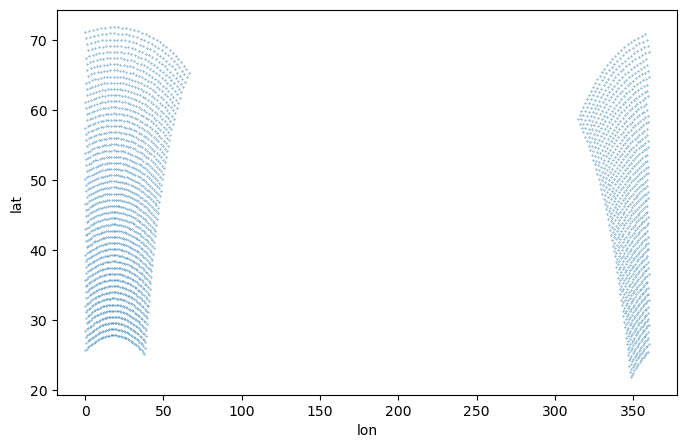
Remap to regional regular lat-lon grid of adaptive resolution with the bilinear method
[9]:
ds_remap = clops.regrid(ds_cordex, method="bilinear", grid="adaptive", output_type="xarray")[0]
ds_remap
[9]:
<xarray.Dataset>
Dimensions: (lat: 201, lon: 225, bnds: 2, time: 1)
Coordinates:
* lat (lat) float64 21.89 22.14 22.39 22.64 ... 71.12 71.37 71.62 71.87
* lon (lon) float64 -45.38 -44.88 -44.38 -43.88 ... 65.18 65.67 66.17
lat_bnds (lat, bnds) float64 21.76 22.01 22.01 22.26 ... 71.75 71.75 72.0
lon_bnds (lon, bnds) float64 -45.62 -45.13 -45.13 ... 65.92 65.92 66.42
height float64 2.0
* time (time) datetime64[ns] 2021-01-16T12:00:00
time_bnds (time, bnds) datetime64[ns] 2021-01-01 2021-02-01
Dimensions without coordinates: bnds
Data variables:
tas (time, lat, lon) float32 nan nan nan nan nan ... nan nan nan nan
Attributes: (12/40)
institution: Helmholtz-Zentrum Geesthacht, Climate Ser...
institute_id: GERICS
experiment_id: rcp85
source: GERICS-REMO2015
model_id: GERICS-REMO2015
forcing: N/A
... ...
NCO: netCDF Operators version 4.7.5 (Homepage ...
grid: regional regular_lat_lon 201x225 (45225 c...
grid_label: gr
regrid_operation: bilinear_201x225_201x225
regrid_tool: xESMF_v0.8.2
regrid_weights_uid: 05603f6f046b266ba6bc164c5b3209bf_cbfbd94f...Plot the remapped data next to the source data
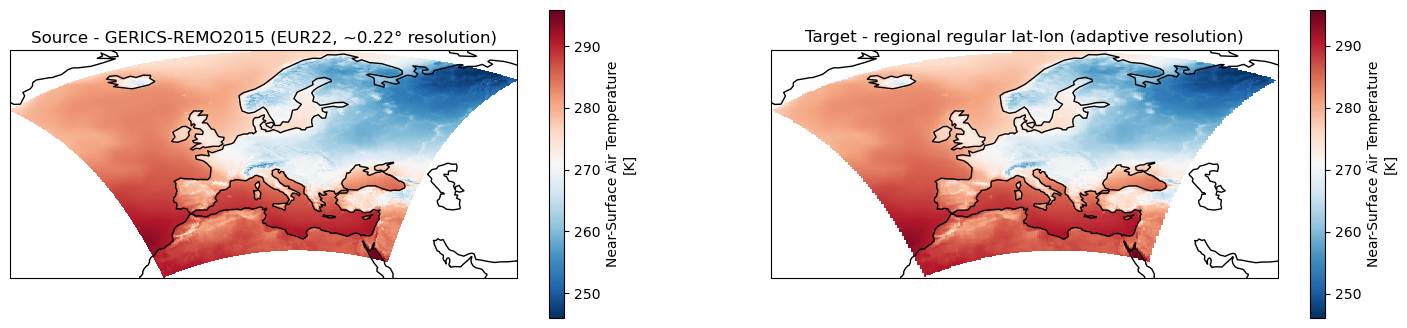
[10]:
fig, axes = plt.subplots(ncols=2, figsize=(18,4), subplot_kw={'projection': ccrs.PlateCarree()})
for ax in axes: ax.coastlines()
# Source data
ds_cordex.tas.isel(time=0).plot.pcolormesh(ax=axes[0], x="lon", y="lat", shading="auto", cmap="RdBu_r")
axes[0].title.set_text("Source - GERICS-REMO2015 (EUR22, ~0.22° resolution)")
# Remapped data
ds_remap.tas.isel(time=0).plot.pcolormesh(ax=axes[1], x="lon", y="lat", shading="auto", cmap="RdBu_r")
axes[1].title.set_text("Target - regional regular lat-lon (adaptive resolution)")
Remap unstructured xarray.Dataset to a global grid of adaptive resolution using the nearest neighbour method
For unstructured grids, at least for the moment, only the nearest neighbour remapping method is supported.
Load the dataset
[11]:
ds_icono_path = Path(MINIESGF, "badc/cmip6/data/CMIP6/CMIP/MPI-M/ICON-ESM-LR/historical/"
"r1i1p1f1/Omon/thetao/gn/v20210215/"
"thetao_Omon_ICON-ESM-LR_historical_r1i1p1f1_gn_185001.nc")
ds_icono = xr.open_dataset(ds_icono_path)
ds_icono
[11]:
<xarray.Dataset>
Dimensions: (time: 1, bnds: 2, i: 235403, vertices: 3, lev: 2)
Coordinates:
* time (time) datetime64[ns] 1850-01-16T12:00:00
longitude (i) float64 ...
latitude (i) float64 ...
* lev (lev) float64 6.0 17.0
Dimensions without coordinates: bnds, i, vertices
Data variables:
time_bnds (time, bnds) datetime64[ns] ...
longitude_bnds (i, vertices) float64 ...
latitude_bnds (i, vertices) float64 ...
lev_bnds (lev, bnds) float64 ...
thetao (time, lev, i) float32 ...
Attributes: (12/51)
CDI: Climate Data Interface version 1.9.8 (https://mpi...
history: Tue Oct 26 11:15:49 2021: cdo sellevidx,1/2 -selt...
source: ICON-ESM-LR (2017): \naerosol: none, prescribed M...
institution: Max Planck Institute for Meteorology
Conventions: CF-1.7 CMIP-6.2
CDI_grid_type: unstructured
... ...
variable_id: thetao
variant_label: r1i1p1f1
license: CMIP6 model data produced by MPI-M is licensed un...
cmor_version: 3.6.0
tracking_id: hdl:21.14100/5e828f18-2231-443c-b560-b1e4ff2a8ef9
CDO: Climate Data Operators version 1.9.8 (https://mpi...Take a look at the grid
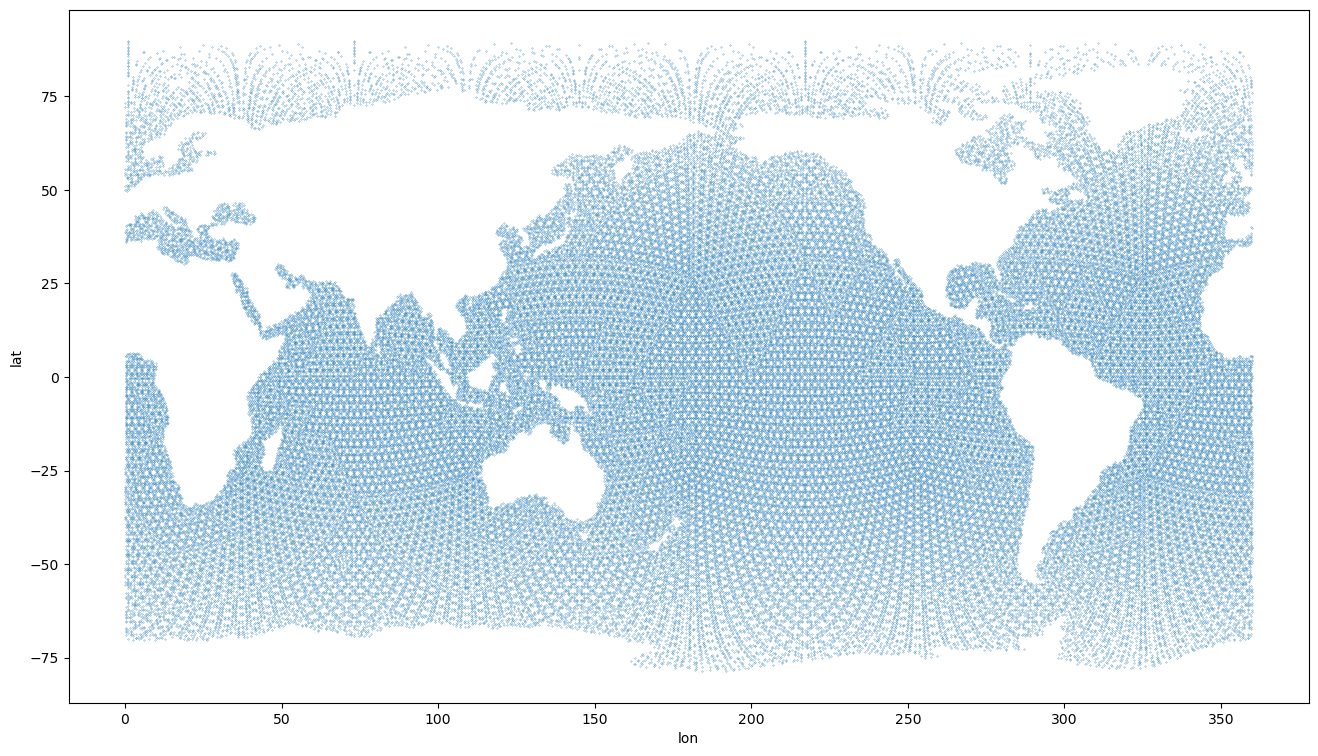
[12]:
plt.figure(figsize=(16,9))
plt.scatter(ds_icono['longitude'][::2], ds_icono['latitude'][::2], s=0.05)
plt.xlabel('lon')
plt.ylabel('lat')
[12]:
Text(0, 0.5, 'lat')
Remap to global grid of adaptive resolution with the nearest neighbour method
[13]:
ds_remap = clops.regrid(ds_icono, method="nearest_s2d", grid="adaptive", output_type="xarray")[0]
ds_remap
[13]:
<xarray.Dataset>
Dimensions: (lat: 331, lon: 709, bnds: 2, time: 1, lev: 2)
Coordinates:
* lat (lat) float64 -78.62 -78.11 -77.6 -77.09 ... 88.7 89.21 89.72
* lon (lon) float64 0.003196 0.5117 1.02 1.529 ... 359.0 359.5 360.0
lat_bnds (lat, bnds) float64 -78.87 -78.36 -78.36 ... 89.46 89.46 89.97
lon_bnds (lon, bnds) float64 -0.251 0.2574 0.2574 ... 359.7 359.7 360.3
* time (time) datetime64[ns] 1850-01-16T12:00:00
* lev (lev) float64 6.0 17.0
time_bnds (time, bnds) datetime64[ns] 1850-01-01 1850-02-01
lev_bnds (lev, bnds) float64 0.0 11.5 11.5 22.0
Dimensions without coordinates: bnds
Data variables:
thetao (time, lev, lat, lon) float32 nan nan nan nan ... -1.8 -1.8 -1.8
Attributes: (12/56)
CDI: Climate Data Interface version 1.9.8 (https...
history: Tue Oct 26 11:15:49 2021: cdo sellevidx,1/2...
source: ICON-ESM-LR (2017): \naerosol: none, prescr...
institution: Max Planck Institute for Meteorology
Conventions: CF-1.7 CMIP-6.2
CDI_grid_type: unstructured
... ...
grid_original: gn
grid_label_original: gn
nominal_resolution_original: 50 km
regrid_operation: nearest_s2d_1x235403_331x709
regrid_tool: xESMF_v0.8.2
regrid_weights_uid: ec7db0ee9226e085bfe8207b8de0672d_e2c8b52440...Plot source data and remapped data
(Using psyplot to plot the unstructured data since xarray does not (yet?) support it.)
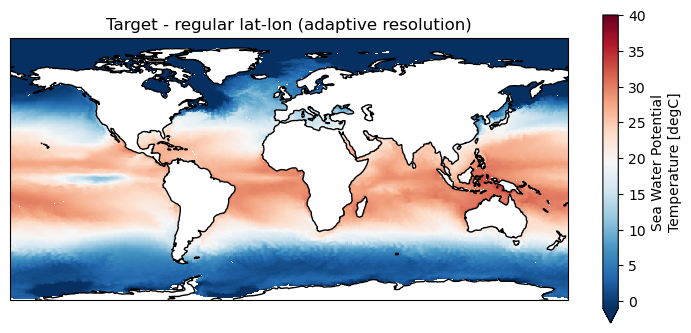
[14]:
# Source data
maps=psy.plot.mapplot(ds_icono_path, cmap="RdBu_r", title="Source - ICON-ESM-LR ICON-O (Ruby-0, 40km resolution)",
time=[0], lev=[0])
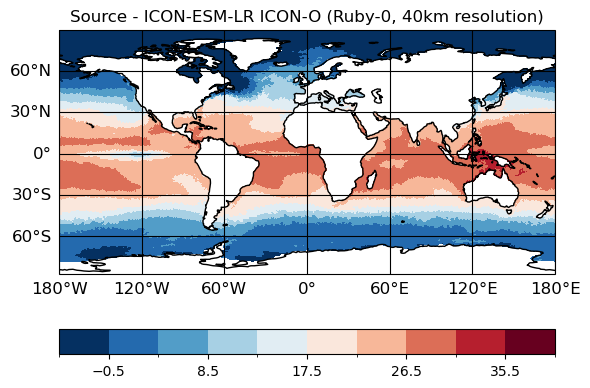
[15]:
# Remapped data
plt.figure(figsize=(9,4));
ax = plt.axes(projection=ccrs.PlateCarree());
ds_remap.thetao.isel(time=0, lev=0).plot.pcolormesh(ax=ax, x="lon", y="lat", shading="auto",
cmap="RdBu_r", vmin = -1, vmax=40)
ax.title.set_text("Target - regular lat-lon (adaptive resolution)")
ax.coastlines()
[15]:
<cartopy.mpl.feature_artist.FeatureArtist at 0x7fe20faa1b10>
clisops.core.Grid
Create a grid object from an xarray.Dataset
Load the dataset
[16]:
dso_path = Path(MINIESGF, "badc/cmip6/data/CMIP6/CMIP/MPI-M/MPI-ESM1-2-HR/historical/r1i1p1f1/Omon/tos/gn/"
"v20190710/tos_Omon_MPI-ESM1-2-HR_historical_r1i1p1f1_gn_185001.nc")
dso = xr.open_dataset(dso_path)
dso
[16]:
<xarray.Dataset>
Dimensions: (i: 802, j: 404, time: 1, bnds: 2, vertices: 4)
Coordinates:
* i (i) int32 0 1 2 3 4 5 6 ... 795 796 797 798 799 800 801
* j (j) int32 0 1 2 3 4 5 6 ... 397 398 399 400 401 402 403
latitude (j, i) float64 ...
longitude (j, i) float64 ...
* time (time) datetime64[ns] 1850-01-16T12:00:00
Dimensions without coordinates: bnds, vertices
Data variables:
time_bnds (time, bnds) datetime64[ns] ...
tos (time, j, i) float32 ...
vertices_latitude (j, i, vertices) float64 ...
vertices_longitude (j, i, vertices) float64 ...
Attributes: (12/48)
Conventions: CF-1.7 CMIP-6.2
activity_id: CMIP
branch_method: standard
branch_time_in_child: 0.0
branch_time_in_parent: 0.0
contact: cmip6-mpi-esm@dkrz.de
... ...
variable_id: tos
variant_label: r1i1p1f1
license: CMIP6 model data produced by MPI-M is licensed un...
cmor_version: 3.5.0
tracking_id: hdl:21.14100/ccd8388f-4f80-4a1e-ba47-66fe65bbeece
NCO: netCDF Operators version 4.7.8 (Homepage = http:/...Create the Grid object
[17]:
grido = clore.Grid(ds=dso)
grido
[17]:
clisops 404x802_cells_grid
Lat x Lon: 404 x 802
Gridcells: 324008
Format: CF
Type: curvilinear
Extent: global
Source: Dataset
Bounds? True
Collapsed cells? True
Duplicated cells? True
Permanent Mask: None
md5 hash: a55c642a80a2b5a8adcc71a9db4fc5f5
The xarray.Dataset is attached to the clisops.core.Grid object. Auxiliary coordinates and data variables have been (re)set appropriately.
[18]:
grido.ds
[18]:
<xarray.Dataset>
Dimensions: (i: 802, j: 404, time: 1, bnds: 2, vertices: 4)
Coordinates:
* i (i) int32 0 1 2 3 4 5 6 ... 795 796 797 798 799 800 801
* j (j) int32 0 1 2 3 4 5 6 ... 397 398 399 400 401 402 403
latitude (j, i) float64 51.15 51.15 51.15 ... -78.67 -78.67
longitude (j, i) float64 83.1 83.06 83.01 ... 82.4 82.85 83.3
* time (time) datetime64[ns] 1850-01-16T12:00:00
time_bnds (time, bnds) datetime64[ns] ...
vertices_latitude (j, i, vertices) float64 51.49 51.39 ... -78.67 -78.48
vertices_longitude (j, i, vertices) float64 83.01 83.0 ... 83.08 83.08
Dimensions without coordinates: bnds, vertices
Data variables:
tos (time, j, i) float32 ...
Attributes: (12/48)
Conventions: CF-1.7 CMIP-6.2
activity_id: CMIP
branch_method: standard
branch_time_in_child: 0.0
branch_time_in_parent: 0.0
contact: cmip6-mpi-esm@dkrz.de
... ...
variable_id: tos
variant_label: r1i1p1f1
license: CMIP6 model data produced by MPI-M is licensed un...
cmor_version: 3.5.0
tracking_id: hdl:21.14100/ccd8388f-4f80-4a1e-ba47-66fe65bbeece
NCO: netCDF Operators version 4.7.8 (Homepage = http:/...Plot the data
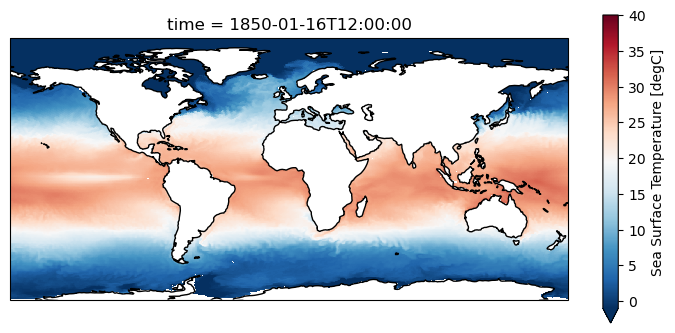
[19]:
plt.figure(figsize=(9,4))
ax = plt.axes(projection=ccrs.PlateCarree())
grido.ds.tos.isel(time=0).plot.pcolormesh(ax=ax, x=grido.lon, y=grido.lat, shading="auto",
cmap="RdBu_r", vmin = -1, vmax=40)
ax.coastlines()
[19]:
<cartopy.mpl.feature_artist.FeatureArtist at 0x7fe20fb23290>
Create a grid object from an xarray.DataArray
Note that xarray.DataArray objects do not support the bounds of coordinate variables to be defined.
Extract tos DataArray
[20]:
dao = dso.tos
dao
[20]:
<xarray.DataArray 'tos' (time: 1, j: 404, i: 802)>
[324008 values with dtype=float32]
Coordinates:
* i (i) int32 0 1 2 3 4 5 6 7 8 ... 794 795 796 797 798 799 800 801
* j (j) int32 0 1 2 3 4 5 6 7 8 ... 396 397 398 399 400 401 402 403
latitude (j, i) float64 51.15 51.15 51.15 51.15 ... -78.67 -78.67 -78.67
longitude (j, i) float64 83.1 83.06 83.01 82.96 ... 81.95 82.4 82.85 83.3
* time (time) datetime64[ns] 1850-01-16T12:00:00
Attributes:
standard_name: sea_surface_temperature
long_name: Sea Surface Temperature
comment: Temperature of upper boundary of the liquid ocean, includ...
units: degC
original_name: tos
cell_methods: area: mean where sea time: mean
cell_measures: area: areacello
history: 2019-08-25T06:00:45Z altered by CMOR: replaced missing va...Create Grid object for MPIOM tos dataarray:
[21]:
grido_tos = clore.Grid(ds=dao)
grido_tos
[21]:
clisops 404x802_cells_grid
Lat x Lon: 404 x 802
Gridcells: 324008
Format: CF
Type: curvilinear
Extent: global
Source: Dataset
Bounds? False
Collapsed cells? None
Duplicated cells? True
Permanent Mask: None
md5 hash: d6214b2150fd961e02d2842ce3533eec
Create a grid object using a grid_instructor
global grid:
grid_instructor = (lon_step, lat_step)orgrid_instructor = stepregional grid:
grid_instructor = (lon_start, lon_end, lon_step, lat_start, lat_end, lat_step)orgrid_instructor = (start, end, step)
[22]:
grid_1deg = clore.Grid(grid_instructor=1)
grid_1deg
[22]:
clisops 180x360_cells_grid
Lat x Lon: 180 x 360
Gridcells: 64800
Format: CF
Type: regular_lat_lon
Extent: global
Source: xESMF
Bounds? True
Collapsed cells? False
Duplicated cells? False
Permanent Mask: None
md5 hash: ff485fa62d5f72e2db23980e3c76efa7
[23]:
grid_1degx2deg_regional = clore.Grid(grid_instructor=(0., 90., 1., 35., 50., 2. ))
grid_1degx2deg_regional
[23]:
clisops 7x90_cells_grid
Lat x Lon: 7 x 90
Gridcells: 630
Format: CF
Type: regular_lat_lon
Extent: regional
Source: xESMF
Bounds? True
Collapsed cells? False
Duplicated cells? False
Permanent Mask: None
md5 hash: c2ddd1634879f48dfa1bdde053198fcb
Create a grid object using a grid_id
Makes use of the predefined grids of roocs_grids, which is a collection of grids used for example for the IPCC Atlas and for CMIP6 Regridding Weights generation.
[24]:
for key, gridinfo in grid_annotations.items(): print(f"- {key:20} {gridinfo}")
- 0pt25deg Global 0.25 degree grid with one cell centered at 0.125E,0.125N
- World_Ocean_Atlas Global 1.0 degree grid with one cell centered at 0.5E,0.5N. As used by the World Ocean Atlas.
- 1deg Global 1.0 degree grid with one cell centered at 0.5E,0.5N. As used by the World Ocean Atlas.
- 2pt5deg Global 2.5 degree grid with one cell centered at 1.25E,1.25N.
- MERRA-2 Global 0.65x0.5 (latxlon) degree grid with one cell centered at 0E,0N. As used by MERRA-2.
- 0pt625x0pt5deg Global 0.65x0.5 (latxlon) degree grid with one cell centered at 0E,0N. As used by MERRA-2.
- ERA-Interim Global 0.75 degree grid with one cell centered at 0E,0N. As used by ERA-Interim.
- 0pt75deg Global 0.75 degree grid with one cell centered at 0E,0N. As used by ERA-Interim.
- ERA-40 Global 1.25 degree grid with one cell centered at 0E,0N. As used by ERA-40.
- 1pt25deg Global 1.25 degree grid with one cell centered at 0E,0N. As used by ERA-40.
- ERA5 Global 0.25 degree grid with one cell centered at 0E,0N. As used by ERA-5.
- 0pt25deg_era5 Global 0.25 degree grid with one cell centered at 0E,0N. As used by ERA-5.
- 0pt25deg_era5_lsm Global 0.25 degree grid with one cell centered at 0E,0N. As used by ERA-5. Includes a fractional land-sea mask.
- 0pt5deg_lsm Global 0.5 degree grid with one cell centered at 0.25E,0.25N. Includes a fractional land-sea mask.
- 1deg_lsm Global 1.0 degree grid with one cell centered at 0.5E,0.5N. As used by the World Ocean Atlas. Includes a fractional land-sea mask.
- 2deg_lsm Global 2.0 degree grid with one cell centered at 1.0E,1.0N.
- 0pt25deg_era5_lsm_binary Global 0.25 degree grid with one cell centered at 0E,0N. As used by ERA-5. Includes a binary land-sea mask with land/sea fraction cut at >=0.5.
- 1deg_lsm_binary Global 1.0 degree grid with one cell centered at 0.5E,0.5N. As used by the World Ocean Atlas. Includes a binary land-sea mask with land/sea fraction cut at >=0.5.
- 2deg_lsm_binary Global 2.0 degree grid with one cell centered at 1.0E,1.0N. Includes a binary land-sea mask with land/sea fraction cut at >=0.5.
- T31 Gaussian global grid of approx. 3.8 degree resolution, 48x96 nlatxnlon. Associated to a T31 spectral grid representation.
- T42 Gaussian global grid of approx. 2.8 degree resolution, 64x128 nlatxnlon. Associated to a T42 spectral grid representation.
- T63_lsm_binary Gaussian global grid of approx. 1.9 degree resolution, 96x192 nlatxnlon. Associated to a T63 spectral grid representation. Includes a binary land-sea mask.
- T127_lsm_binary Gaussian global grid of approx. 1.0 degree resolution, 192x384 nlatxnlon. Associated to a T127 spectral grid representation. Includes a binary land-sea mask.
- T255 Gaussian global grid of approx. 0.5 degree resolution, 384x768 nlatxnlon. Associated to a T255 spectral grid representation.
[25]:
grid_era5 = clore.Grid(grid_id = "0pt25deg_era5")
grid_era5
[25]:
clisops 721x1440_cells_grid
Lat x Lon: 721 x 1440
Gridcells: 1038240
Format: CF
Type: regular_lat_lon
Extent: global
Source: Predefined_0pt25deg_era5
Bounds? True
Collapsed cells? False
Duplicated cells? False
Permanent Mask: None
md5 hash: cdf4f59eab828857bc0c2819c0c465d7
clisops.core.Grid objects can be compared to one another
Optional verbose output gives information on where the grids differ: lat, lon, lat_bnds, lon_bnds, mask?
Compare the tos dataset to the tos dataarray
[26]:
comp = grido.compare_grid(grido_tos, verbose = True)
print("Grids are equal?", comp)
The two grids differ in their respective lat_bnds, lon_bnds.
Grids are equal? False
Compare both 0.25° ERA5 Grids
[27]:
# Create the Grid object
grid_era5_lsm = clore.Grid(grid_id = "0pt25deg_era5_lsm", compute_bounds=True)
[28]:
# Compare
comp = grid_era5.compare_grid(grid_era5_lsm, verbose=True)
print("Grids are equal?", comp)
The two grids are considered equal.
Grids are equal? True
clisops.core.Weights
Create regridding weights to regrid between two grids. Supported are the following of xESMF’s remapping methods: * nearest_s2d * bilinear * conservative * patch
Create 2-degree target grid
[34]:
grid_2deg = clore.Grid(grid_id="2deg_lsm", compute_bounds=True)
grid_2deg
[34]:
clisops 90x180_cells_grid
Lat x Lon: 90 x 180
Gridcells: 16200
Format: CF
Type: regular_lat_lon
Extent: global
Source: Predefined_2deg_lsm
Bounds? True
Collapsed cells? False
Duplicated cells? False
Permanent Mask: None
md5 hash: 438f79b4376004d360a56fa2c0abb9f0
Create conservative remapping weights using the clisops.core.Weights class
grid_in and grid_out are Grid objects
[35]:
%time weights = clore.Weights(grid_in = grido, grid_out = grid_2deg, method="conservative")
CPU times: user 10.8 s, sys: 344 ms, total: 11.2 s
Wall time: 11.2 s
Local weights cache
Weights are cached on disk and do not have to be created more than once. The default cache directory is platform-dependent and set via the package platformdirs. For Linux it is '/home/my_user/.local/share/clisops/weights_dir' and can optionally be adjusted:
permanently by modifying the parameter
grid_weights: local_weights_dirin theroocs.iniconfiguration file that can be found in the clisops installation directoryor temporarily via:
from clisops import core as clore
clore.weights_cache_init("/dir/for/weights/cache")
[36]:
from clisops.core.regrid import CONFIG
print(CONFIG["clisops:grid_weights"]["local_weights_dir"])
/home/docs/.local/share/clisops/grid_weights
[37]:
!ls -sh {CONFIG["clisops:grid_weights"]["local_weights_dir"]}
total 32M
904K grid_05603f6f046b266ba6bc164c5b3209bf.nc
32K grid_20d38455170b1db562edb4f5e06d3cc6.nc
24K grid_438f79b4376004d360a56fa2c0abb9f0.nc
8.6M grid_a55c642a80a2b5a8adcc71a9db4fc5f5.nc
24K grid_cbfbd94fc4feee0c6b34841124e4adf8.nc
20K grid_d4ebf553238ebf865140bded4c8e73b9.nc
40K grid_e2c8b52440eb2392c600702aa1c532ef.nc
7.3M grid_ec7db0ee9226e085bfe8207b8de0672d.nc
4.0K weights_05603f6f046b266ba6bc164c5b3209bf_cbfbd94fc4feee0c6b34841124e4adf8_unperi_no-degen_bilinear.json
2.0M weights_05603f6f046b266ba6bc164c5b3209bf_cbfbd94fc4feee0c6b34841124e4adf8_unperi_no-degen_bilinear.nc
4.0K weights_20d38455170b1db562edb4f5e06d3cc6_d4ebf553238ebf865140bded4c8e73b9_peri_no-degen_bilinear.json
1.5M weights_20d38455170b1db562edb4f5e06d3cc6_d4ebf553238ebf865140bded4c8e73b9_peri_no-degen_bilinear.nc
4.0K weights_a55c642a80a2b5a8adcc71a9db4fc5f5_438f79b4376004d360a56fa2c0abb9f0_peri_skip-degen_conservative.json
7.6M weights_a55c642a80a2b5a8adcc71a9db4fc5f5_438f79b4376004d360a56fa2c0abb9f0_peri_skip-degen_conservative.nc
4.0K weights_ec7db0ee9226e085bfe8207b8de0672d_e2c8b52440eb2392c600702aa1c532ef_peri_no-degen_nearest_s2d.json
3.6M weights_ec7db0ee9226e085bfe8207b8de0672d_e2c8b52440eb2392c600702aa1c532ef_peri_no-degen_nearest_s2d.nc
[38]:
!cat {CONFIG["clisops:grid_weights"]["local_weights_dir"]}/weights_*_conservative.json
{
"def_filename": "conservative_404x802_90x180.nc",
"filename": "weights_a55c642a80a2b5a8adcc71a9db4fc5f5_438f79b4376004d360a56fa2c0abb9f0_peri_skip-degen_conservative.nc",
"format": "xESMF",
"ignore_degenerate": "True",
"method": "conservative",
"periodic": "True",
"source_extent": "global",
"source_format": "CF",
"source_lat": "latitude",
"source_lat_bnds": "vertices_latitude",
"source_lon": "longitude",
"source_lon_bnds": "vertices_longitude",
"source_ncells": 324008,
"source_nlat": 404,
"source_nlon": 802,
"source_source": "/home/docs/.mini-esgf-data/master/test_data/badc/cmip6/data/CMIP6/CMIP/MPI-M/MPI-ESM1-2-HR/historical/r1i1p1f1/Omon/tos/gn/v20190710/tos_Omon_MPI-ESM1-2-HR_historical_r1i1p1f1_gn_185001.nc",
"source_tracking_id": "hdl:21.14100/ccd8388f-4f80-4a1e-ba47-66fe65bbeece",
"source_type": "curvilinear",
"source_uid": "a55c642a80a2b5a8adcc71a9db4fc5f5",
"target_extent": "global",
"target_format": "CF",
"target_lat": "lat",
"target_lat_bnds": "lat_bnds",
"target_lon": "lon",
"target_lon_bnds": "lon_bnds",
"target_ncells": 16200,
"target_nlat": 90,
"target_nlon": 180,
"target_source": "/home/docs/checkouts/readthedocs.org/user_builds/clisops/conda/stable/lib/python3.11/site-packages/roocs_grids/grids/land_sea_mask_2degree.nc4",
"target_tracking_id": "",
"target_type": "regular_lat_lon",
"target_uid": "438f79b4376004d360a56fa2c0abb9f0",
"tool": "xESMF_v0.8.2",
"uid": "a55c642a80a2b5a8adcc71a9db4fc5f5_438f79b4376004d360a56fa2c0abb9f0_peri_skip-degen_conservative"
}
Now the weights will be read directly from the cache
[39]:
%time weights = clore.Weights(grid_in = grido, grid_out = grid_2deg, method="conservative")
CPU times: user 223 ms, sys: 32 ms, total: 255 ms
Wall time: 255 ms
The weights cache can be flushed, which removes all weight and grid files as well as the json files holding the metadata. To see what would be removed, one can use the dryrun=True parameter. To re-initialize the weights cache in a different directory, one can use the weights_dir_init="/new/dir/for/weights/cache" parameter. Even when re-initializing the weights cache under a new path, using clore.weights_cache_flush, no directory is getting removed, only above listed files. When
dryrun is not set, the files that are getting deleted can be displayed with verbose=True.
[40]:
clore.weights_cache_flush(dryrun=True)
Flushing the clisops weights cache ('/home/docs/.local/share/clisops/grid_weights') would remove:
- /home/docs/.local/share/clisops/grid_weights/weights_20d38455170b1db562edb4f5e06d3cc6_d4ebf553238ebf865140bded4c8e73b9_peri_no-degen_bilinear.json
- /home/docs/.local/share/clisops/grid_weights/weights_05603f6f046b266ba6bc164c5b3209bf_cbfbd94fc4feee0c6b34841124e4adf8_unperi_no-degen_bilinear.json
- /home/docs/.local/share/clisops/grid_weights/weights_ec7db0ee9226e085bfe8207b8de0672d_e2c8b52440eb2392c600702aa1c532ef_peri_no-degen_nearest_s2d.json
- /home/docs/.local/share/clisops/grid_weights/weights_a55c642a80a2b5a8adcc71a9db4fc5f5_438f79b4376004d360a56fa2c0abb9f0_peri_skip-degen_conservative.json
- /home/docs/.local/share/clisops/grid_weights/weights_20d38455170b1db562edb4f5e06d3cc6_d4ebf553238ebf865140bded4c8e73b9_peri_no-degen_bilinear.nc
- /home/docs/.local/share/clisops/grid_weights/weights_a55c642a80a2b5a8adcc71a9db4fc5f5_438f79b4376004d360a56fa2c0abb9f0_peri_skip-degen_conservative.nc
- /home/docs/.local/share/clisops/grid_weights/weights_05603f6f046b266ba6bc164c5b3209bf_cbfbd94fc4feee0c6b34841124e4adf8_unperi_no-degen_bilinear.nc
- /home/docs/.local/share/clisops/grid_weights/weights_ec7db0ee9226e085bfe8207b8de0672d_e2c8b52440eb2392c600702aa1c532ef_peri_no-degen_nearest_s2d.nc
- /home/docs/.local/share/clisops/grid_weights/grid_ec7db0ee9226e085bfe8207b8de0672d.nc
- /home/docs/.local/share/clisops/grid_weights/grid_20d38455170b1db562edb4f5e06d3cc6.nc
- /home/docs/.local/share/clisops/grid_weights/grid_e2c8b52440eb2392c600702aa1c532ef.nc
- /home/docs/.local/share/clisops/grid_weights/grid_a55c642a80a2b5a8adcc71a9db4fc5f5.nc
- /home/docs/.local/share/clisops/grid_weights/grid_d4ebf553238ebf865140bded4c8e73b9.nc
- /home/docs/.local/share/clisops/grid_weights/grid_cbfbd94fc4feee0c6b34841124e4adf8.nc
- /home/docs/.local/share/clisops/grid_weights/grid_438f79b4376004d360a56fa2c0abb9f0.nc
- /home/docs/.local/share/clisops/grid_weights/grid_05603f6f046b266ba6bc164c5b3209bf.nc
[41]:
clore.weights_cache_flush(verbose=True)
Flushing the clisops weights cache ('/home/docs/.local/share/clisops/grid_weights'). Removing ...
- /home/docs/.local/share/clisops/grid_weights/weights_20d38455170b1db562edb4f5e06d3cc6_d4ebf553238ebf865140bded4c8e73b9_peri_no-degen_bilinear.json
- /home/docs/.local/share/clisops/grid_weights/weights_05603f6f046b266ba6bc164c5b3209bf_cbfbd94fc4feee0c6b34841124e4adf8_unperi_no-degen_bilinear.json
- /home/docs/.local/share/clisops/grid_weights/weights_ec7db0ee9226e085bfe8207b8de0672d_e2c8b52440eb2392c600702aa1c532ef_peri_no-degen_nearest_s2d.json
- /home/docs/.local/share/clisops/grid_weights/weights_a55c642a80a2b5a8adcc71a9db4fc5f5_438f79b4376004d360a56fa2c0abb9f0_peri_skip-degen_conservative.json
- /home/docs/.local/share/clisops/grid_weights/weights_20d38455170b1db562edb4f5e06d3cc6_d4ebf553238ebf865140bded4c8e73b9_peri_no-degen_bilinear.nc
- /home/docs/.local/share/clisops/grid_weights/weights_a55c642a80a2b5a8adcc71a9db4fc5f5_438f79b4376004d360a56fa2c0abb9f0_peri_skip-degen_conservative.nc
- /home/docs/.local/share/clisops/grid_weights/weights_05603f6f046b266ba6bc164c5b3209bf_cbfbd94fc4feee0c6b34841124e4adf8_unperi_no-degen_bilinear.nc
- /home/docs/.local/share/clisops/grid_weights/weights_ec7db0ee9226e085bfe8207b8de0672d_e2c8b52440eb2392c600702aa1c532ef_peri_no-degen_nearest_s2d.nc
- /home/docs/.local/share/clisops/grid_weights/grid_ec7db0ee9226e085bfe8207b8de0672d.nc
- /home/docs/.local/share/clisops/grid_weights/grid_20d38455170b1db562edb4f5e06d3cc6.nc
- /home/docs/.local/share/clisops/grid_weights/grid_e2c8b52440eb2392c600702aa1c532ef.nc
- /home/docs/.local/share/clisops/grid_weights/grid_a55c642a80a2b5a8adcc71a9db4fc5f5.nc
- /home/docs/.local/share/clisops/grid_weights/grid_d4ebf553238ebf865140bded4c8e73b9.nc
- /home/docs/.local/share/clisops/grid_weights/grid_cbfbd94fc4feee0c6b34841124e4adf8.nc
- /home/docs/.local/share/clisops/grid_weights/grid_438f79b4376004d360a56fa2c0abb9f0.nc
- /home/docs/.local/share/clisops/grid_weights/grid_05603f6f046b266ba6bc164c5b3209bf.nc
Initialized new weights cache at /home/docs/.local/share/clisops/grid_weights
clisops.core.regrid
This function allows to perform the eventual regridding and provides a resulting xarray.Dataset
def regrid(
grid_in: Grid,
grid_out: Grid,
weights: Weights,
adaptive_masking_threshold: Optional[float] = 0.5,
keep_attrs: Optional[bool] = True,
):
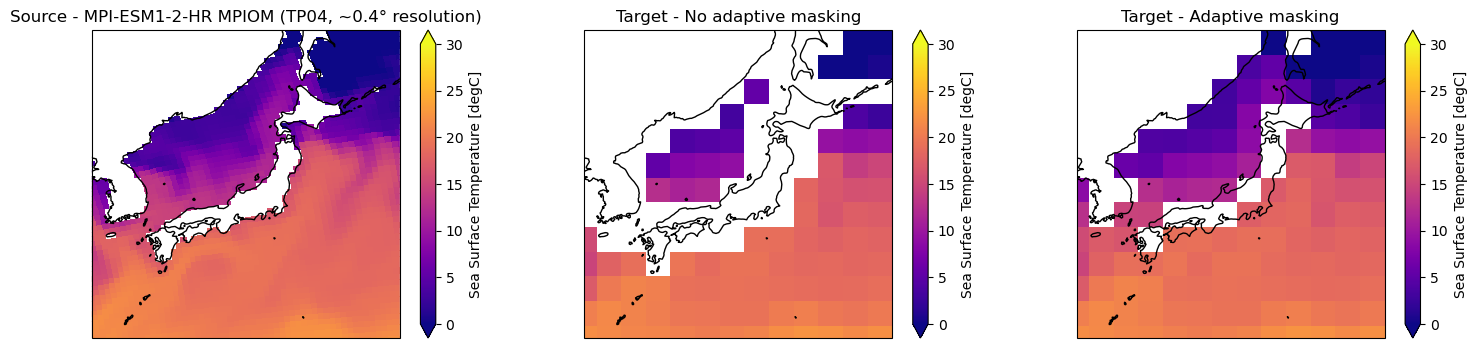
grid_inandgrid_outareGridobjects,weightsis aWeightsobject.adaptive_masking_threshold(AMT) A value within the [0., 1.] interval that defines the maximumRATIOof missing_values amongst the total number of data values contributing to the calculation of the target grid cell value. For a fraction [0., AMT[ of the contributing source data missing, the target grid cell will be set to missing_value, else, it will be re-normalized by the factor1./(1.-RATIO). Thus, if AMT is set to 1, all source grid cells that contribute to a target grid cell must be missing in order for the target grid cell to be defined as missing itself. Values greater than 1 or less than 0 will cause adaptive masking to be turned off. This adaptive masking technique allows to reuse generated weights for differently masked data (e.g. land-sea masks or orographic masks that vary with depth / height).keep_attrscan have the following settings:True: The resultingxarray.Datasetwill have all attributes ofgrid_in.ds.attrs, despite attributes that have to be added and altered due to the new grid.False: The resultingxarray.Datasetwill have no attributes despite attributes generated by the regridding process."target": The resultingxarray.Datasetwill have all attributes ofgrid_out.ds.attrs, despite attributes generated by the regridding process. Not recommended.
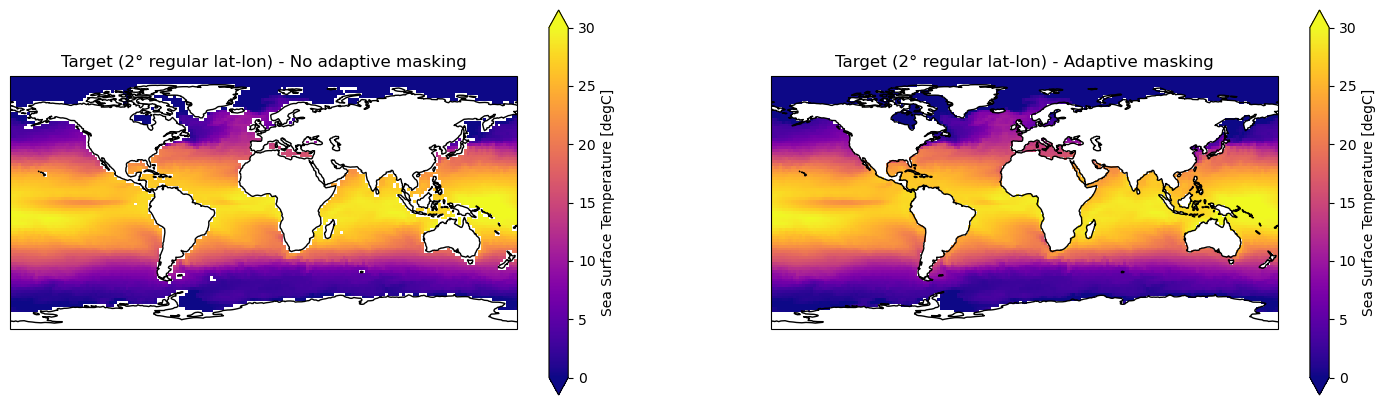
In the following an example showing the function application and the effect of the adaptive masking.
[42]:
ds_out_amt0 = clore.regrid(grido, grid_2deg, weights, adaptive_masking_threshold=-1)
[43]:
ds_out_amt1 = clore.regrid(grido, grid_2deg, weights, adaptive_masking_threshold=0.5)
Plot the resulting data
[44]:
# Create panel plot of regridded data (global)
fig, axes = plt.subplots(ncols=2, nrows=1,
figsize=(18, 5), # global
subplot_kw={'projection': ccrs.PlateCarree()})
ds_out_amt0["tos"].isel(time=0).plot.pcolormesh(ax=axes[0], vmin=0, vmax=30, cmap="plasma")
axes[0].title.set_text("Target (2° regular lat-lon) - No adaptive masking")
ds_out_amt1["tos"].isel(time=0).plot.pcolormesh(ax=axes[1], vmin=0, vmax=30, cmap="plasma")
axes[1].title.set_text("Target (2° regular lat-lon) - Adaptive masking")
for axis in axes.flatten():
axis.coastlines()
axis.set_xlabel('lon')
axis.set_ylabel('lat')
[45]:
# Create panel plot of regridded data (Japan)
fig, axes = plt.subplots(ncols=3, nrows=1,
figsize=(18, 4), # Japan
subplot_kw={'projection': ccrs.PlateCarree()})
grido.ds.tos.isel(time=0).plot.pcolormesh(ax=axes[0], x=grido.lon, y=grido.lat,
vmin=0, vmax=30, cmap="plasma", shading="auto")
axes[0].title.set_text("Source - MPI-ESM1-2-HR MPIOM (TP04, ~0.4° resolution)")
ds_out_amt0["tos"].isel(time=0).plot.pcolormesh(ax=axes[1], vmin=0, vmax=30, cmap="plasma")
axes[1].title.set_text("Target - No adaptive masking")
ds_out_amt1["tos"].isel(time=0).plot.pcolormesh(ax=axes[2], vmin=0, vmax=30, cmap="plasma")
axes[2].title.set_text("Target - Adaptive masking")
for axis in axes.flatten():
axis.coastlines()
axis.set_xlabel('lon')
axis.set_ylabel('lat')
axis.set_xlim([125, 150])
axis.set_ylim([25, 50])